Chromosomes are minute hair strand-like structures that are present inside an animal cell or plant cell.
What chromosomes are made up of- A chromosome consists of certain proteins along with DNA that are neatly aligned on the genes.Chromosomes have a densely coiled DNA molecule which is located inside the nucleus of the majorly all cells like animal cell and plant cell.
This article completely focuses on the what chromosomes are made up of like their components, structure and various other factors.
The chromosome is a portion or region in the nucleus that has the genetic information of the specific organism and aids in the transfer of the genetic information from the parent to young ones.
Read More on Do Bacteria Have Chloroplasts? Why, What Type, How And Detailed Facts
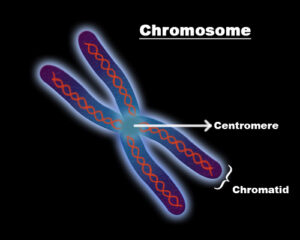
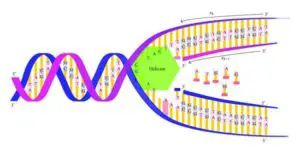
Image credits- Flickr
What are the chromosome components?
The chromosome is a portion or region in the nucleus that has the genetic information of the specific organism. The chromosome components are
- Two Identical chromatids- One is the exact copy of the other and has the DNA molecule.
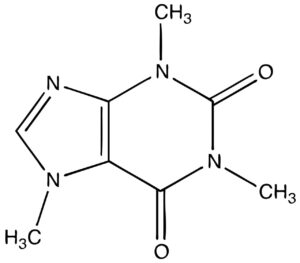
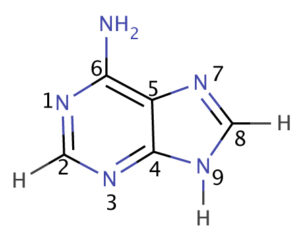
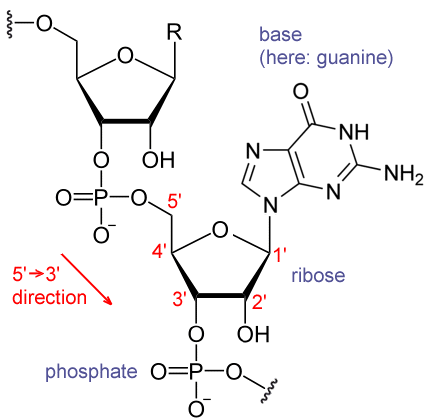
- DNA– A densely packed molecule of DNA that gives the genetic information.
- Protein-The DNA molecule is complexly wrapped or bound with a specific protein which is called the histones.
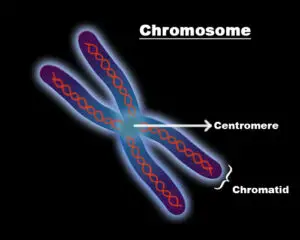
- Centromere– The connective or the middle narrow point of the arms, thai point is called centromere.
- p-Arm- The short arm in the chromosome.
- q-Arm- The long arm in the chromosome.
- Telomere- The end or the terminal point of the chromosome.
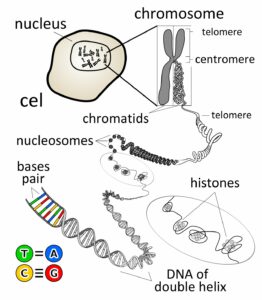
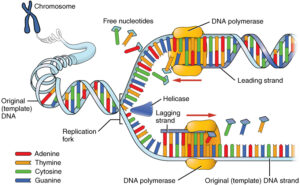
Image credits- Wikimedia
Read more on Biosynthesis of Purines and Pyrimidines | An important part of cellular metabolism
What chromosomes are made up of?
The structure of the chromosome is made up of various components which will be discussed below.
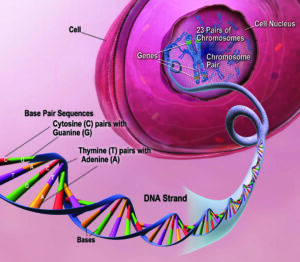
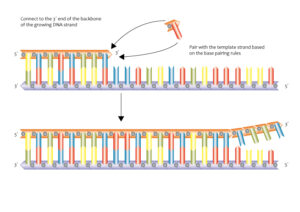
Image credits- Flickr
Read More on Is Fungi Prokaryotic Or Eukaryotic: Why, How And Detailed Insights And Facts
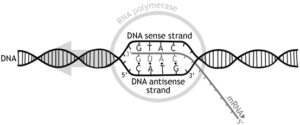
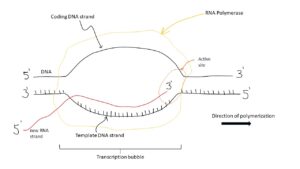
Chromatin:
- Chromatin is a system or network in a chromosome which has the genetic material and the associated protein.
- Chromatin is of 2 formation. One of the 2 formations is called the Euchromatin which is low condensed and the transcription can happen. The other form is called heterochromatin, it is highly condensed and no transcription can happen.
- The chromatin has a bead-like structure called the nucleosome, which is bound with a protein called solenoid that acts as a support.
Read More on Chromatin Organization | Its Crucial impact on the packaging of DNA
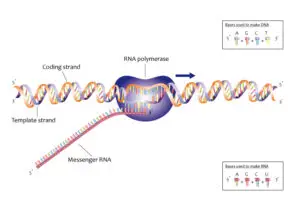
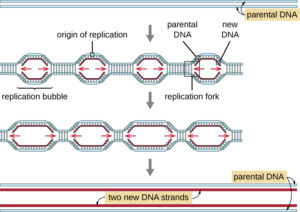
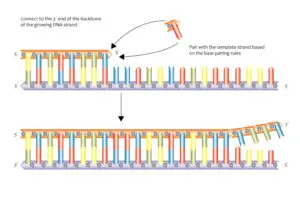
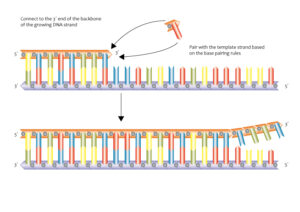
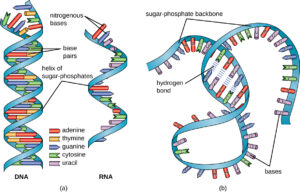
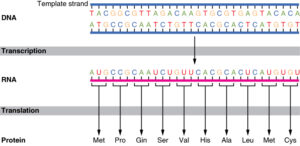
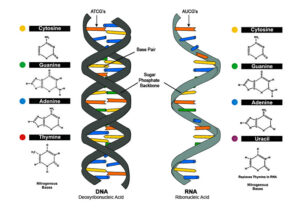
DNA Molecule:
- The genetic material that has the genetic information is present in the DNA molecule.
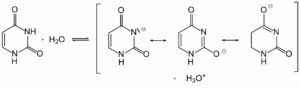
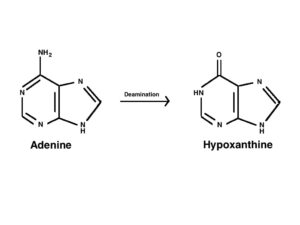
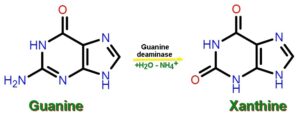
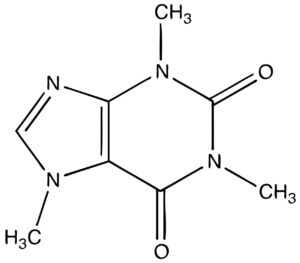
- DNA molecule has the Nitrogenous base (Purine- Adenine and Guanine and Pyrimidine- Thymine, Cytosine), A pentose sugar molecule which is called as Deoxyribose, A highly negative phosphate molecule which is the backbone of the complete structure.
Protein:
The DNA molecule is bound or wrapped with the associated proteins which is called the histone protein that completely form to make nucleosomes.
Read More on Is Fungi Prokaryotic Or Eukaryotic: Why, How And Detailed Insights And Facts
Centromere:
- The narrow point or the constriction point in which the 2 identical chromatids unite or join is called the centromere.
- Centromere is also called the kinetochore and 2 types are usually seen which are the point centromere and regional centromere.
Arm:
Two arms are present
- p-Arm- The short arm in the chromosome.
- q-Arm- The long arm in the chromosome.
Read More on Nucleotide Excision Repair and Single Nucleotide Polymorphism | An Important discussion
Telomere:
- The end point or terminal point of the chromosome is called the telomere.
- It consists of the non repetitive sequence at the end which is held to protect the chromosome from any external damage.
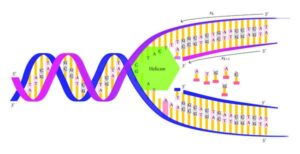
Image credits- Flickr
What are bacterial chromosomes made up of?
- Bacteria have only one chromosome or single chromosome.
- It has a double stranded DNA structure in loop form (circular structure).
- In bacteria, the DNA is present inside the nucleoid of the bacterial cell and does not possess any associated proteins.
- Usually the bacteria have small bits of looped DNA structure called the plasmids that replicate independently.
- The plasmids have less number of genes like around 30 only.
- Eg: In E. Coli- the length of the chromosome when stretched is as long as many times of the length of the same cell.
- They are highly and tightly packed inside the portion of the cell.
Read more on Adenosine nucleoside and nucleoside phosphoramidite | Overview of important aspects
List of organisms and the number of chromosomes present inside them.
- Rice has 24 Number of chromosomes
- Oats has 42 Number of chromosomes
- Wheat has 42 Number of chromosomes
- Mango has 40 Number of chromosomes
- Mold has 4 Number of chromosomes
- Human has 46 Number of chromosomes
- Monkey has 42 Number of chromosomes
- Fruit fly has 8 Number of chromosomes
- Flatworm has 16 Number of chromosomes
- Potato has 48 Number of chromosomes
- Cat has 38 Number of chromosomes
- Lion has 38 Number of chromosomes
- Dog has 78 Number of chromosomes
- Elephant has 58 Number of chromosomes
- Kingfisher has 132 Number of chromosomes
Also Read: