DNA replication is a fundamental process that ensures the accurate duplication of genetic information, enabling the faithful transmission of hereditary traits from one generation to the next. At the heart of this complex mechanism lie a diverse array of enzymes, each playing a crucial role in orchestrating the seamless replication of the DNA double helix. In this comprehensive guide, we will delve into the intricacies of the key DNA replication enzymes, providing a detailed exploration of their functions, kinetics, and error rates.
DNA Polymerases: The Maestros of DNA Synthesis
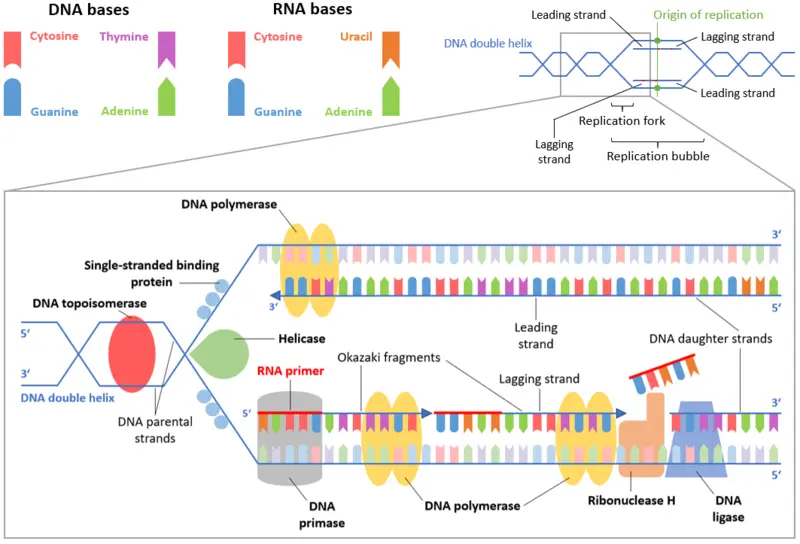
DNA polymerases are the primary enzymes responsible for the synthesis of new DNA strands, adding nucleotides to the growing chain in a 5′ to 3′ direction. While there are several types of DNA polymerases, each with specialized functions, the workhorse of DNA replication is DNA polymerase III.
- DNA Polymerase III: This enzyme is responsible for the bulk of DNA synthesis during replication, with an impressive speed of 500-1000 nucleotides per second in E. coli. Its high fidelity is achieved through a combination of accurate nucleotide selection and a proofreading mechanism that can correct errors at a rate of approximately 1 mistake per 10^4 to 10^6 nucleotides synthesized.
- DNA Polymerase I: This enzyme plays a crucial role in removing RNA primers and replacing them with DNA, ensuring the continuity of the newly synthesized strands. DNA polymerase I has a lower error rate of around 1 mistake per 10^5 to 10^6 nucleotides synthesized.
- DNA Polymerase II: While less well-studied, DNA polymerase II is believed to be involved in DNA repair processes, providing a backup mechanism to ensure the integrity of the genetic material.
DNA Primase: The Primer Provider

DNA primase is a specialized enzyme that synthesizes short RNA primers on the single-stranded DNA template, providing the necessary starting points for DNA polymerase to initiate DNA synthesis. The error rate of DNA primase is approximately 1 mistake per 10^3 to 10^4 nucleotides synthesized, slightly higher than that of the DNA polymerases.
DNA Helicase: The Unwinding Maestro
DNA helicase is the enzyme responsible for separating the two strands of the DNA double helix, creating the replication fork where DNA synthesis can occur. This enzyme moves in a 5′ to 3′ direction, unwinding the DNA as it progresses. The speed of DNA helicase varies among different organisms, with E. coli helicase moving at a rate of 500 to 1000 base pairs per second.
DNA Ligase: The Sealing Specialist
DNA ligase plays a crucial role in sealing the gaps between Okazaki fragments on the lagging strand, forming a continuous DNA strand. This enzyme has an error rate of approximately 1 mistake per 10^5 to 10^6 nucleotides ligated, ensuring a high degree of fidelity in the final DNA product.
Topoisomerase: The Tension Relievers
As the replication fork progresses, the DNA double helix ahead of it can become increasingly tightly wound, creating topological stress that could impede the replication process. Topoisomerase enzymes alleviate this tension by making temporary nicks in the DNA helix, allowing the strands to unwind, and then sealing the nicks to maintain the integrity of the genetic material. The rate of topoisomerase activity varies among different organisms, with E. coli topoisomerase I relaxing supercoiled DNA at a rate of 500 to 1000 base pairs per second.
In addition to these key enzymes, DNA replication also involves a host of other proteins and enzymes, such as single-stranded binding proteins, which coat the single-stranded DNA around the replication fork to prevent rewinding, and primase, which synthesizes the RNA primers complementary to the DNA strand.
The intricate dance of these DNA replication enzymes, each with its unique function and kinetic properties, ensures the accurate and efficient duplication of the genetic code, laying the foundation for the continued evolution and survival of living organisms.
References:
- Lodish, H., Berk, A., Zipursky, S. L., Matsudaira, P., Baltimore, D., & Darnell, J. (2000). Molecular cell biology. Scientific American books.
- Lehninger, A. L., Nelson, D. L., & Cox, M. M. (2000). Lehninger principles of biochemistry. W.H. Freeman.
- Alberts, B., Johnson, A., Lewis, J., Raff, M., Roberts, K., & Walter, P. (2002). Molecular biology of the cell. Garland Science.
- Watson, J. D., Baker, T. A., Bell, S. P., Gann, A., Levine, M., & Losick, R. (2013). Molecular biology of the gene. Pearson.
- Replication of DNA: Enzymes and the Replication Fork. (n.d.). Retrieved from https://www.nature.com/scitable/topicpage/replication-of-dna-enzymes-and-the-replication-408/
- DNA Replication Enzymes. (n.d.). Retrieved from https://www.genome.gov/genetics-glossary/DNA-Replication-Enzymes