Antiparallel DNA strands refer to the arrangement of the two strands in a DNA molecule. In this arrangement, the two strands run in opposite directions, with one strand running in the 5′ to 3′ direction and the other running in the 3′ to 5′ direction. This antiparallel orientation is crucial for DNA replication and transcription processes. The antiparallel arrangement allows for complementary base pairing between the strands, where adenine (A) pairs with thymine (T) and guanine (G) pairs with cytosine (C). This complementary base pairing is essential for maintaining the genetic code and ensuring accurate DNA replication and protein synthesis.
Key Takeaways
| Fact | Description |
|---|---|
| Antiparallel DNA strands | Two DNA strands running in opposite directions |
| Direction of the strands | One strand runs in the 5′ to 3′ direction, while the other runs in the 3′ to 5′ direction |
| Complementary base pairing | Adenine (A) pairs with thymine (T), and guanine (G) pairs with cytosine (C) |
| Importance in DNA processes | Crucial for DNA replication and transcription |
| Maintenance of genetic code | Ensures accurate DNA replication and protein synthesis |
Understanding the Structure of DNA
DNA, short for deoxyribonucleic acid, is a molecule that carries the genetic instructions for the development, functioning, and reproduction of all living organisms. It is often referred to as the “blueprint of life.” Understanding the structure of DNA is crucial in the field of molecular biology as it provides insights into how genetic information is stored and transmitted.
The Antiparallel Nature of DNA Strands
One of the key features of DNA is its antiparallel nature. This means that the two strands of DNA run in opposite directions. One strand runs in the 5′ to 3′ direction, while the other runs in the 3′ to 5′ direction. This antiparallel arrangement is essential for DNA replication and the accurate transmission of genetic information.
Structural Features of DNA
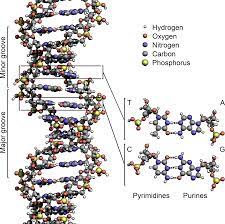
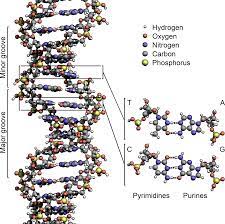
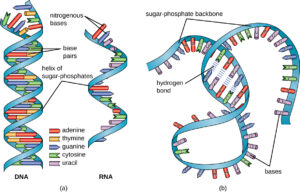
DNA has a double helix structure, resembling a twisted ladder. This structure is formed by two complementary strands of nucleotides held together by hydrogen bonds. The nucleotides consist of a sugar molecule (deoxyribose), a phosphate group, and one of four nitrogenous bases: adenine (A), thymine (T), cytosine (C), and guanine (G).
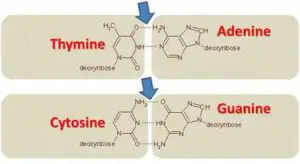
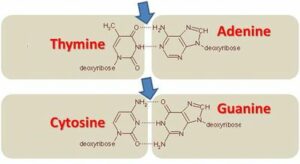
The base pairing between the nitrogenous bases is specific and follows the rules of complementary base pairing. Adenine always pairs with thymine, forming two hydrogen bonds, while cytosine always pairs with guanine, forming three hydrogen bonds. This base pairing ensures the stability and integrity of the DNA molecule.
The Watson-Crick model, proposed by James Watson and Francis Crick in 1953, provided the first accurate description of the DNA structure. According to this model, the two DNA strands are twisted around each other in a helical fashion, with the nitrogenous bases facing inward. This arrangement allows for efficient base pairing and easy access to the genetic information.
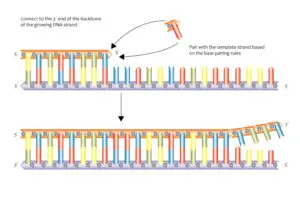
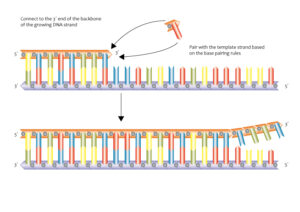
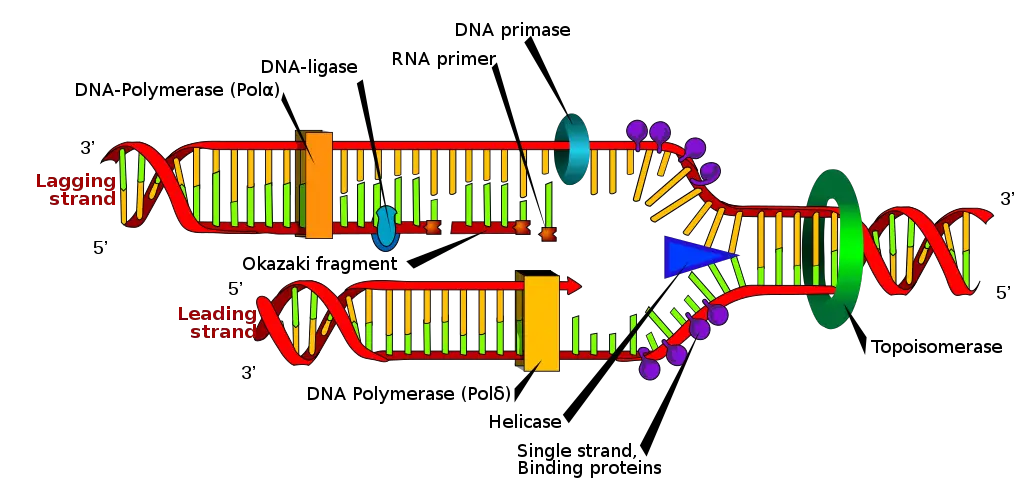
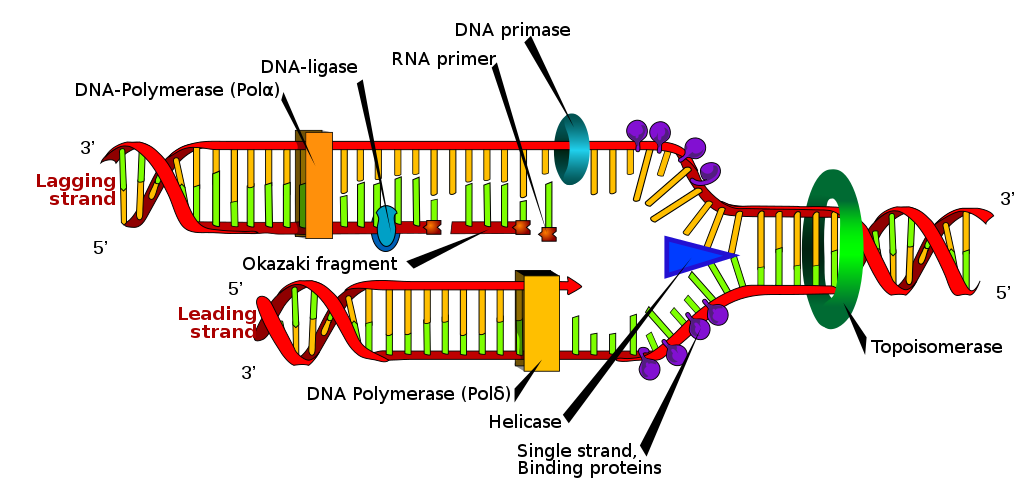
DNA replication is a fundamental process in which the DNA molecule is duplicated. It occurs during cell division and is essential for the transmission of genetic information to daughter cells. The replication process involves the action of various enzymes, including DNA polymerase, DNA helicase, and DNA ligase.
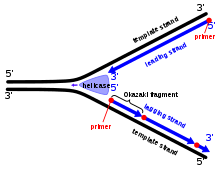
During replication, the DNA strands separate at specific sites called replication forks. DNA helicase unwinds the double helix, creating two single strands. DNA polymerase then adds complementary nucleotides to each single strand, following the 5′ to 3′ direction. The leading strand is synthesized continuously, while the lagging strand is synthesized in short fragments called Okazaki fragments. These fragments are later joined by DNA ligase to form a continuous strand.
DNA topology refers to the three-dimensional arrangement of DNA in space. DNA supercoiling is a common phenomenon in which the DNA molecule becomes twisted upon itself. DNA gyrase is an enzyme that helps relieve the tension caused by supercoiling, ensuring the proper functioning of DNA.
Understanding the structure of DNA and its various features is crucial for unraveling the mysteries of the genetic code. It provides insights into how genetic information is stored, replicated, and transmitted, paving the way for advancements in fields such as medicine, agriculture, and biotechnology.
The Antiparallel Arrangement of DNA Strands
The antiparallel arrangement of DNA strands is a fundamental characteristic of the double helix structure, which is the iconic shape of DNA. This arrangement refers to the orientation of the two strands running in opposite directions. In other words, while one strand runs from the 5′ to 3′ direction, the other strand runs from the 3′ to 5′ direction. This antiparallel arrangement plays a crucial role in DNA replication and other essential cellular processes.
How are DNA Strands Antiparallel?
The antiparallel arrangement of DNA strands is a consequence of the base pairing between nucleotides. Each nucleotide consists of a sugar molecule, a phosphate group, and a nitrogenous base. The two strands of DNA are held together by hydrogen bonds formed between complementary base pairs. Adenine (A) pairs with thymine (T), and guanine (G) pairs with cytosine (C).
In the Watson-Crick model of DNA, the two strands are oriented in opposite directions. One strand has its 5′ end (containing the phosphate group) at the top, while the other strand has its 3′ end at the top. This arrangement allows for the complementary base pairing to occur, ensuring the stability and integrity of the DNA molecule.
Why are DNA Strands Antiparallel?
The antiparallel arrangement of DNA strands is essential for DNA replication. During replication, the DNA molecule unwinds at specific sites called replication forks. DNA helicase enzymes separate the two strands by breaking the hydrogen bonds between the base pairs. As the strands separate, DNA polymerase enzymes synthesize new strands by adding complementary nucleotides.
The antiparallel arrangement allows DNA polymerase to synthesize new strands in the 5′ to 3′ direction. The leading strand is synthesized continuously in the 5′ to 3′ direction, following the replication fork. However, the lagging strand is synthesized discontinuously in small fragments called Okazaki fragments. These fragments are later joined together by DNA ligase.
The antiparallel arrangement also plays a role in DNA topology and supercoiling. DNA gyrase enzymes help relieve the tension caused by the unwinding of the DNA molecule during replication. They introduce negative supercoils by temporarily breaking and rejoining the DNA strands.
In summary, the antiparallel arrangement of DNA strands is a crucial aspect of the double helix structure. It facilitates base pairing, DNA replication, and other essential processes in molecular biology. Understanding this arrangement helps us unravel the mysteries of the genetic code and the intricate workings of life itself.
The Significance of Antiparallel DNA Strands
Antiparallel DNA strands play a crucial role in various biological processes, particularly in DNA replication and genetic information transfer. Understanding the significance of antiparallel DNA strands is essential for comprehending the intricate mechanisms that govern molecular biology.
Role in DNA Replication
During DNA replication, the double helix structure of DNA unwinds to expose the two complementary strands. The antiparallel nature of these strands is vital for the accurate replication of genetic material.
DNA replication occurs in a 5′ to 3′ direction, meaning that the new DNA strand is synthesized in the opposite direction to the parental template strand. The antiparallel arrangement of the DNA strands allows for the continuous synthesis of one strand, known as the leading strand, while the other strand, called the lagging strand, is synthesized in short fragments known as Okazaki fragments.
The process of DNA replication involves several key enzymes and proteins. DNA helicase unwinds the double helix, creating a replication fork where the two strands separate. DNA polymerase then adds nucleotides to the growing DNA strand, following the complementary base pairing rules. The antiparallel arrangement ensures that the DNA polymerase can synthesize the new strand in the correct direction.
To join the Okazaki fragments on the lagging strand, DNA ligase plays a crucial role. It seals the gaps between the fragments, resulting in a continuous DNA strand. The antiparallel nature of the DNA strands is essential for the proper functioning of DNA ligase and the seamless completion of DNA replication.
Importance in Genetic Information Transfer
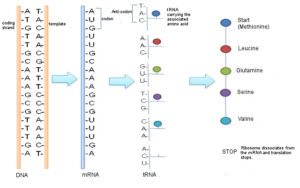
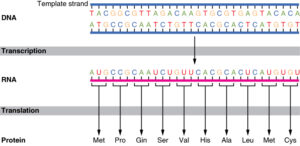
Antiparallel DNA strands also play a vital role in the transfer of genetic information. The complementary base pairing between the two strands allows for the accurate transmission of the genetic code during processes such as transcription and translation.
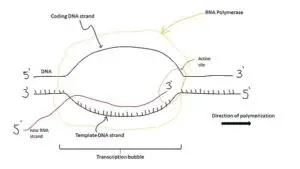
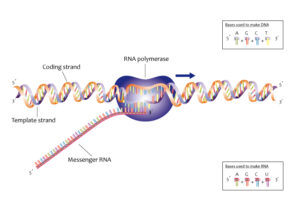
During transcription, the DNA sequence is transcribed into RNA by RNA polymerase. The antiparallel arrangement ensures that the RNA molecule is synthesized in a complementary manner to the DNA template strand. This process allows for the faithful transfer of genetic information from DNA to RNA.
In translation, the mRNA molecule is used as a template to synthesize proteins. The antiparallel nature of DNA strands ensures that the mRNA sequence is complementary to the DNA template strand, allowing for the correct translation of the genetic code into amino acids.
Furthermore, the antiparallel arrangement of DNA strands also plays a role in DNA topology and supercoiling. DNA gyrase, an enzyme involved in DNA topology, helps relieve the torsional strain caused by the winding of DNA strands. The antiparallel arrangement allows DNA gyrase to efficiently manage the supercoiling of DNA, ensuring the proper functioning of the genetic material.
In conclusion, the significance of antiparallel DNA strands cannot be overstated in molecular biology. From DNA replication to genetic information transfer, the antiparallel arrangement ensures the accurate transmission and faithful replication of the genetic code. Understanding the role of antiparallel DNA strands provides valuable insights into the fundamental processes that govern life itself.
Exploring the Antiparallel DNA Structure
The double helix structure of DNA is a fundamental concept in molecular biology. It consists of two complementary strands that run in opposite directions, known as antiparallel strands. In this article, we will delve into the intricacies of the antiparallel DNA structure, including the labeling of antiparallel DNA strands and the potential disadvantages associated with them.
Labeling Antiparallel DNA Strands

Labeling antiparallel DNA strands is crucial for understanding the structure and function of DNA. The labeling process involves identifying the orientation of the strands and determining the 5′ to 3′ directionality. The 5′ end of a DNA strand has a phosphate group attached to the 5th carbon of the sugar molecule, while the 3′ end has a hydroxyl group attached to the 3rd carbon. By labeling the strands, scientists can analyze the nucleotide sequence and study the interactions between the strands.
To label antiparallel DNA strands, researchers often use fluorescent dyes or radioactive isotopes. These labels allow for visualization and tracking of the DNA strands during experiments. Additionally, specific techniques such as DNA sequencing rely on accurate labeling to determine the order of nucleotides in a DNA molecule.
Potential Disadvantages of Antiparallel DNA Strands
While the antiparallel DNA structure is essential for DNA replication and other cellular processes, it does present some potential disadvantages. One such disadvantage is the formation of DNA knots and tangles due to the topological properties of the antiparallel strands. DNA topology refers to the different ways in which DNA can be twisted, coiled, or knotted.
During DNA replication, the unwinding of the double helix by DNA helicase creates a replication fork. As the replication fork moves along the DNA molecule, the antiparallel strands can become tangled, leading to the formation of knots. These knots can impede the progress of DNA replication and cause errors in the genetic code.
To overcome these knots and tangles, cells employ enzymes like DNA gyrase, which can introduce temporary breaks in the DNA strands and relieve the tension. Another mechanism involves the synthesis of short DNA fragments, known as Okazaki fragments, on the lagging strand during DNA replication. These fragments are later joined together by DNA ligase.
In conclusion, the antiparallel DNA structure plays a crucial role in maintaining the integrity and stability of the genetic material. By labeling the antiparallel strands and understanding their potential disadvantages, scientists can gain valuable insights into the intricate workings of DNA and its role in molecular biology.
Frequently Asked Questions
Does DNA have Antiparallel Strands?
Yes, DNA does have antiparallel strands. This means that the two strands of DNA run in opposite directions. One strand runs in the 5′ to 3′ direction, while the other runs in the 3′ to 5′ direction. The antiparallel nature of DNA is an essential feature of its structure.
Why do DNA Strands need to be Antiparallel?
The antiparallel arrangement of DNA strands is crucial for several reasons. Firstly, it allows for the formation of the double helix structure. The complementary strands of DNA are held together by hydrogen bonds between the nitrogenous bases. The antiparallel arrangement ensures that the bases can pair up correctly, with adenine (A) always pairing with thymine (T) and guanine (G) always pairing with cytosine (C).
Secondly, the antiparallel nature of DNA is essential for DNA replication. During replication, the DNA strands separate, and each strand serves as a template for the synthesis of a new complementary strand. The 5′ to 3′ directionality of one strand allows for continuous replication, while the 3′ to 5′ directionality of the other strand leads to the formation of short Okazaki fragments on the lagging strand.
What does it mean that DNA is Antiparallel?
When we say that DNA is antiparallel, we mean that the two strands of the DNA molecule run in opposite directions. The 5′ end of one strand is aligned with the 3′ end of the other strand. This arrangement is known as the Watson-Crick model, named after the scientists who proposed the structure of DNA.
The antiparallel nature of DNA is crucial for the functioning of enzymes involved in DNA replication, such as DNA polymerase. These enzymes can only add nucleotides to the 3′ end of a growing DNA strand. Therefore, the antiparallel arrangement ensures that DNA replication can occur smoothly and accurately.
In addition to DNA replication, the antiparallel strands of DNA also play a role in DNA topology and supercoiling. The winding and twisting of DNA strands can result in the formation of knots and tangles. Enzymes like DNA gyrase help relieve these topological stresses by introducing temporary breaks in the DNA strands and allowing them to rotate.
In summary, the antiparallel nature of DNA strands is a fundamental aspect of DNA’s structure and function. It enables base pairing, DNA replication, and proper DNA topology. Understanding the antiparallel arrangement is essential for studying molecular biology and deciphering the genetic code.
Conclusion
In conclusion, antiparallel DNA strands play a crucial role in the structure and function of DNA. The antiparallel arrangement of the two strands allows for efficient replication and transcription processes. The complementary base pairing between the strands ensures the accurate transmission of genetic information during DNA replication and protein synthesis. Additionally, the antiparallel orientation of the strands contributes to the stability and integrity of the DNA molecule. Understanding the concept of antiparallel DNA strands is essential in comprehending the fundamental mechanisms of genetics and molecular biology.
References
In the field of molecular biology, understanding the structure and function of DNA is crucial. The discovery of the double helix structure by Watson and Crick in 1953 revolutionized our understanding of genetics and paved the way for further research in the field. This groundbreaking model explained how DNA’s base pairing and complementary strands allow for accurate DNA replication and the transmission of genetic information.
To comprehend the intricate process of DNA replication, it is essential to grasp the concept of the 5′ to 3′ direction. DNA replication occurs in this specific direction, where new nucleotides are added to the growing DNA strand. The nucleotide sequence is precisely maintained through base pairing, where adenine (A) pairs with thymine (T) and cytosine (C) pairs with guanine (G) through hydrogen bonds.
The Watson-Crick model of DNA replication elucidates the role of various enzymes in this process. DNA polymerase is responsible for synthesizing new DNA strands by adding nucleotides to the existing template strands. This enzyme ensures the accuracy of DNA replication by proofreading and correcting any errors that may occur.
During DNA replication, the double helix unwinds at specific sites called replication forks. DNA helicase plays a crucial role in this unwinding process by breaking the hydrogen bonds between the complementary strands. As the replication fork progresses, the leading strand is synthesized continuously, while the lagging strand is synthesized in short fragments known as Okazaki fragments.
To connect these Okazaki fragments and complete the synthesis of the lagging strand, an enzyme called DNA ligase is required. This enzyme seals the gaps between the fragments, resulting in a continuous DNA strand. Additionally, primer DNA is necessary to initiate DNA replication, providing a starting point for DNA polymerase to begin synthesizing new DNA strands.
DNA topology and supercoiling also play significant roles in DNA replication. DNA gyrase is an enzyme that helps relieve the tension and strain caused by the unwinding of the DNA double helix during replication. It achieves this by introducing temporary breaks in the DNA strands, allowing them to unwind and prevent the formation of knots or tangles.
In conclusion, the process of DNA replication is a complex and highly regulated mechanism that ensures the accurate transmission of genetic information. Understanding the key players, such as DNA polymerase, DNA helicase, DNA ligase, and the role of DNA topology, is essential in unraveling the mysteries of the genetic code and advancing our knowledge in the field of molecular biology.
What are the Detailed Explanations of the 5 Coliform Bacteria Examples?
Coliform bacteria explained in detail refers to the thorough explanations of the five examples of these bacteria. These include Escherichia coli, Enterobacter aerogenes, Klebsiella pneumoniae, Citrobacter freundii, and Enterobacter cloacae. Each bacterium has its own distinct characteristics, habitats, and potential health implications. Understanding these details helps in identifying and managing their presence in water, food, and other environments.
[]
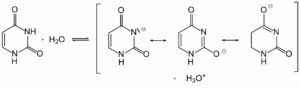
Are antiparallel DNA strands and the pyrimidine nature of adenine connected?
Adenine, one of the four nucleobases found in DNA molecules, has long been recognized as a purine base. However, recent research has shed light on its pyrimidine nature as well. Adenine’s classification as both a purine and a pyrimidine base stems from its ability to form two different tautomeric forms. This discovery has interesting implications, particularly when considering the arrangement of DNA strands. The antiparallel orientation of DNA strands, where one strand runs in the opposite direction to its complementary strand, may interact with adenine’s pyrimidine nature in intriguing ways. To gain a deeper understanding of how these concepts intersect, delve into the article Adenine: Understanding Its Pyrimidine Nature.
Frequently Asked Questions
Q1: Does DNA have antiparallel strands?
Yes, DNA does have antiparallel strands. In the double helix structure of DNA, the two strands run in opposite directions, one from 5′ to 3′ and the other from 3′ to 5′. This is what is referred to as antiparallel.
Q2: What is the significance of antiparallel DNA strands?
The antiparallel nature of DNA strands is crucial for DNA replication. It allows the DNA polymerase to add nucleotides to the 3′ end of the new strand, ensuring accurate and efficient replication of the genetic code.
Q3: What is meant by the term ‘antiparallel structure of DNA strands’?
The term ‘antiparallel structure of DNA strands’ refers to the orientation of the two strands in a DNA molecule. One strand runs in the 5′ to 3′ direction, while the other runs in the 3′ to 5′ direction. This is a key feature of the double helix structure of DNA.
Q4: Why are the two DNA strands antiparallel?
The two DNA strands are antiparallel to facilitate the process of DNA replication. This orientation allows enzymes like DNA polymerase and DNA helicase to function properly, ensuring the accurate copying of the genetic code.
Q5: What does it mean when we say ‘DNA is antiparallel’?
When we say ‘DNA is antiparallel’, we are referring to the orientation of the two strands in a DNA molecule. In the double helix structure, one strand runs from 5′ to 3′ and the other runs from 3′ to 5′. This antiparallel arrangement is crucial for processes like DNA replication and transcription.
Q6: What are the ‘antiparallel strands of DNA’?
The ‘antiparallel strands of DNA’ refer to the two complementary strands in a DNA molecule that run in opposite directions. This means one strand runs from 5′ to 3′ and the other from 3′ to 5′. These strands are held together by hydrogen bonds between complementary base pairs.
Q7: Why do DNA strands need to be antiparallel?
DNA strands need to be antiparallel for the process of DNA replication to occur. The enzymes involved in DNA replication, such as DNA polymerase and DNA helicase, require this antiparallel structure to function correctly.
Q8: How does the antiparallel nature affect DNA replication?
The antiparallel nature of DNA strands affects DNA replication by dictating the direction in which the new strands are synthesized. DNA polymerase can only add nucleotides to the 3′ end of the new strand, resulting in one strand (the leading strand) being synthesized continuously, while the other (the lagging strand) is synthesized in fragments, known as Okazaki fragments.
Q9: What is the role of antiparallel DNA strands in the Watson-Crick model?
In the Watson-Crick model of DNA, the antiparallel strands form the double helix structure. The strands are held together by hydrogen bonds between complementary base pairs, with one strand running from 5′ to 3′ and the other from 3′ to 5′. This model explains how DNA replicates and how the genetic code is preserved.
Q10: Why are DNA strands antiparallel to each other?
DNA strands are antiparallel to each other to ensure accurate DNA replication. This orientation allows the enzymes involved in replication to function properly, ensuring the genetic code is accurately copied and passed on to the next generation.
Also Read: