The process in which the mRNA molecule is synthesized from a DNA strand, it is called Transcription.
DNA transcription process is a complicated mechanism in which a DNA portion is converted or copied into a RNA molecule. The central dogma is a complicated process in which the DNA is transformed into a messenger RNA molecule and the messenger RNA is translated into a protein molecule.
Read more on Biosynthesis of Purines and Pyrimidines | An important part of cellular metabolism
What is the DNA transcription Process?
Every cell in every organism needs to be repaired and rejuvenated at one point of time.
DNA transcription is a process in which a segment of the DNA molecule is transcribed or converted or copied into a RNA molecule to be more precise the DNA molecule get copied to a messenger RNA molecule.
This is called the DNA transcription process.
DNA (segment) gets converted to a messenger RNA molecule.
Read More on Is Fungi Prokaryotic Or Eukaryotic: Why, How And Detailed Insights And Facts
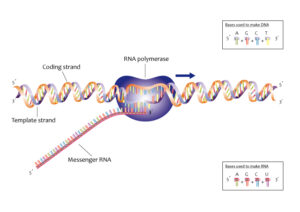
Image credits- Wikimedia
What is the DNA transcription process in prokaryotic organisms?
The DNA transcription process in prokaryotes is quite simple and it is of 3 stages or steps.
DNA transcription process is a complicated mechanism in which a DNA portion is converted or copied into a RNA molecule-Messenger RNA with the help of an enzyme called RNA polymerase.
Initiation of the DNA transcription process:
- Initiation is a process in which the process starts.
- This process is initiated at a particular segment of DNA sequence which is commonly called the promoter region.
- The enzyme RNA polymerase incorporates Ribo- nucleotides to the template DNA strand.
Initiation consists have 3 types
Image credits- Flickr
Closed complex
- Closed complex is a stage in which the RNA polymerase adheres or bind to the promoter region of a DNA sequence.
- Like in DNA replication which uses the enzyme DNA polymerase, RNA polymerase does not need any enzyme like primase to initiate its desired process.
Open complex
- Open complex is the stage when the enzyme that is held together to the DNA sequence unwinds or release the double stranded DNA into a single strand at a certain or particular portion, this is called the open complex.
- The incorporation of rNTPs or the ribo-nucleotides to the template DNA will be done.
- The final product will be short stretches of RNA strand produced complementary to the template DNA.
Abortive initiation
- Abortive initiation is a process in which the sigma factors inhibit the RNA exit channel of RNA polymerase enzymes.
Elongation of the DNA transcription process:
- The sigma factor in the RNA polymerase will be freed from the complex and the elongation of the RNA strand complementary to the template DNA strand will be performed.
- Elongationis a process in which the biosynthesis of an RNA strand happens,the strand will be elongating by the addition of rNTPs or ribo- nucleotides.
Termination of the DNA transcription process:
There has to be an end point to end this transcription process and this is called the termination process or the end process.
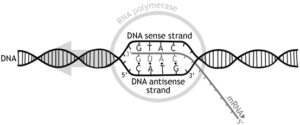
Image credits- Wikimedia
The prokaryotic termination process will be of 2 types.
Rho independent:
- The other name of the Rho independent is intrinsic terminators.
- This kind of termination does not need any external proteins.
This consist of 2 sequence elements, they are
Short inverted repeats
Here are few examples of inverted repeats
Example – I
– – – – – – – – G T A G C A T T C G G – – – – – – – – – C C G A A T G C T A C – – – – – –
– – – – – – – – C A T C G T A A G C C – – – – – – – – – G G C T T A C G A T G – – – – – – –
Example – II
– – – – – – – – T A G C A T T C G G T- – – – – – – – A C C G A A T G C T A- – – – – – –
– – – – – – – – A T C G T A A G C C A- – – – – – – – T G G C T T A C G A T- – – – – – –
Example – III
– – – – – – – – G T A G C A T T C G G T- – – – – – – – A C C G A A T G C T A C- – – – – – –
– – – – – – – – C A T C G T A A G C C A- – – – – – – -T G G C T T A C G A T G- – – – – – –
So from the above examples, we can see that in the 2 strands of the DNA the nitrogenous bases are repeated in the inversion form on its complementary strand with minimal gaps (having other nitrogenous bases).
A T rich sequence
– – G T A G C A T T C G G – – – – – – – – C C G A A T G C T A C – – – – A A A A
– – C A T C G T A A G C C – – – – – – – – G G C T T A C G A T G – – – – T T T T
- A T rich sequence is a sequence in which continuously the strand has adenine that complements thymine or vice versa.
- First the inverted repeated sequences will be present and then they are followed by AT rich sequences.
- Now due to this inverted sequence and AT rich sequence at the terminal point, a hairpin or inverted U shape like structure will be produced by base pairing in the same strand which then stops the complete process.
- Then A T rich sequence becomes A U rich sequence as in RNA thymine is replaced by uracil.
- Among all the base pairs ( A – T ; T – A ; C – G ; G – C ; A – U; U – A), the weakest pair will be A-U or U – A.
- Due to this weak pairing element, this is the terminal point and the RNA will be released or freed and the process is terminated.
Rho dependent:
- The Rho dependent termination process will need a Rho protein to end the process of transcription.
- This Rho protein is a hexameric protein and needs Adenosine triphosphate molecules for its action.
- This Rho protein adheres to the RNA sequence which has more number of cytosine or C, which are also known as Rho utilization sites or the RUT site.
– – – – – – – – C C C C C C – – – – – – –
- This will eventually become the Rho sensitive pause site and unwinds the region.
- Now the final product will be a strand of RNA that is let out from the template DNA, this is the process of prokaryotic DNA transcription.
Read more on Adenosine nucleoside and nucleoside phosphoramidite | Overview of important aspects
What is the DNA transcription process in eukaryotic organisms?
The transcription process is quite complex in eukaryotic organisms.
The eukaryotic organism transcription has 3 steps: Initiation, elongation and termination.
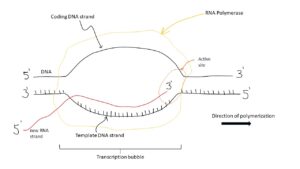
Image credits- Wikimedia
Initiation of the eukaryotic transcription process:
- In this process, 3 RNA polymerase (RNA polymerase I,RNA polymerase II,RNA polymerase III) is used. More importantly RNA polymerase II contributes more in the eukaryotic transcription process.
- There are few transcription factors involved which are called the TF.
Read more on Is Fungi Multicellular Or Unicellular: Why, How And Detailed Insights And Facts
The types of TFs are
The eukaryotic promoter is about 40 nucleotides long located in the upstream and downstream in the sequence.
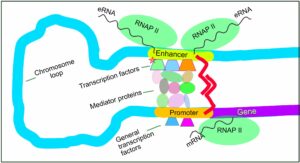
Image credits- Wikimedia
TFIID :
- Initially, the transcription factor TFIID binds to the TATA element of the promoter.
- TFIID produces a protein called the TATA binding protein OR TBP and this protein binds to the TATA sequence resulting in the bending of the sequences to about 80 degree.
- This bending helps in the adhesion of other factors like TFIIA and TFIIB.
TFIIA :
TFIIA aids in stabilizing the binding of TFIID into the promoter sequences.
TFIIB :
- TFIIB interacts with TBP or the TATA binding proteins at the promoter region downstream to the sequence.
- The TFIIB aids in the recruitment of the RNA polymerase II on the promoter as the RNA polymerase can not bind on its own.
TFIIF :
TFIIF aids in the binding of RNA polymerase II into the promoter sequence.
TFIIE :
TFIIE helps to start the preinitiation complex.
TFIIH
- TFIIH is a large complex consisting of 9 subunits, 2 subunits have ATPase activity which provides energy and it acts like helicase and unwinds the DNA.
- This results- From preinitiation complex to open complex.
The other 7 subunits acts as a kinase activity and binds to the RNA polymerase tail and moves to the next step elongation.
Read more on Nucleotide Excision Repair and Single Nucleotide Polymorphism | An Important discussion
Elongation of the eukaryotic transcription process:
- The elongation factors are the components that aid in the elongation process.
- TFEB is a kinase protein and undergoes phosphorylation which stimulates elongation and TFIIS is like a catalyst which aids in the fastening of the elongation process by not letting the RNA polymerase to pause.
- 5′ capping is a process in which certain enzymes add and delete few compounds that aid in the initiation of the translation process.
Termination of the eukaryotic transcription process:
- The RNA polymerase reaches the end point, 2 proteins called the CstF and CPSF interact with the transcription complex and cleaves the messenger RNA from the complex.
- Every component from the complex dissociates.
- CPSF then adds or recruits poly A polymerase which adds about 200 bases of adenine at the 3 ‘ end resulting in the poly A tail by utilizing ATP and the process stops.
Also Read:
- Obligate bacteria examples
- Coconut crab characteristics
- Is cytosine a pyrimidine
- Endoplasmic reticulum in cytoplasm
- Mycorrhizal fungi examples
- Does transcription occur in the nucleus
- Specialist species examples
- Mantis shrimp types
- Are endonucleases restriction enzymes
- Spoilage bacteria examples

Hello, I am Sugaprabha Prasath, a Postgraduate in the field of Microbiology. I am an active member of the Indian association of applied microbiology (IAAM). I have research experience in preclinical (Zebrafish), bacterial enzymology, and nanotechnology. I have published 2 research articles in an International journal and a few more are yet to be published, 2 sequences were submitted to NCBI-GENBANK. I am good at clearly explaining the concepts in biology at both basic and advanced levels. My area of specialization is biotechnology, microbiology, enzymology, molecular biology, and pharmacovigilance. Apart from academics, I love gardening and being with plants and animals.
My LinkedIn profile-