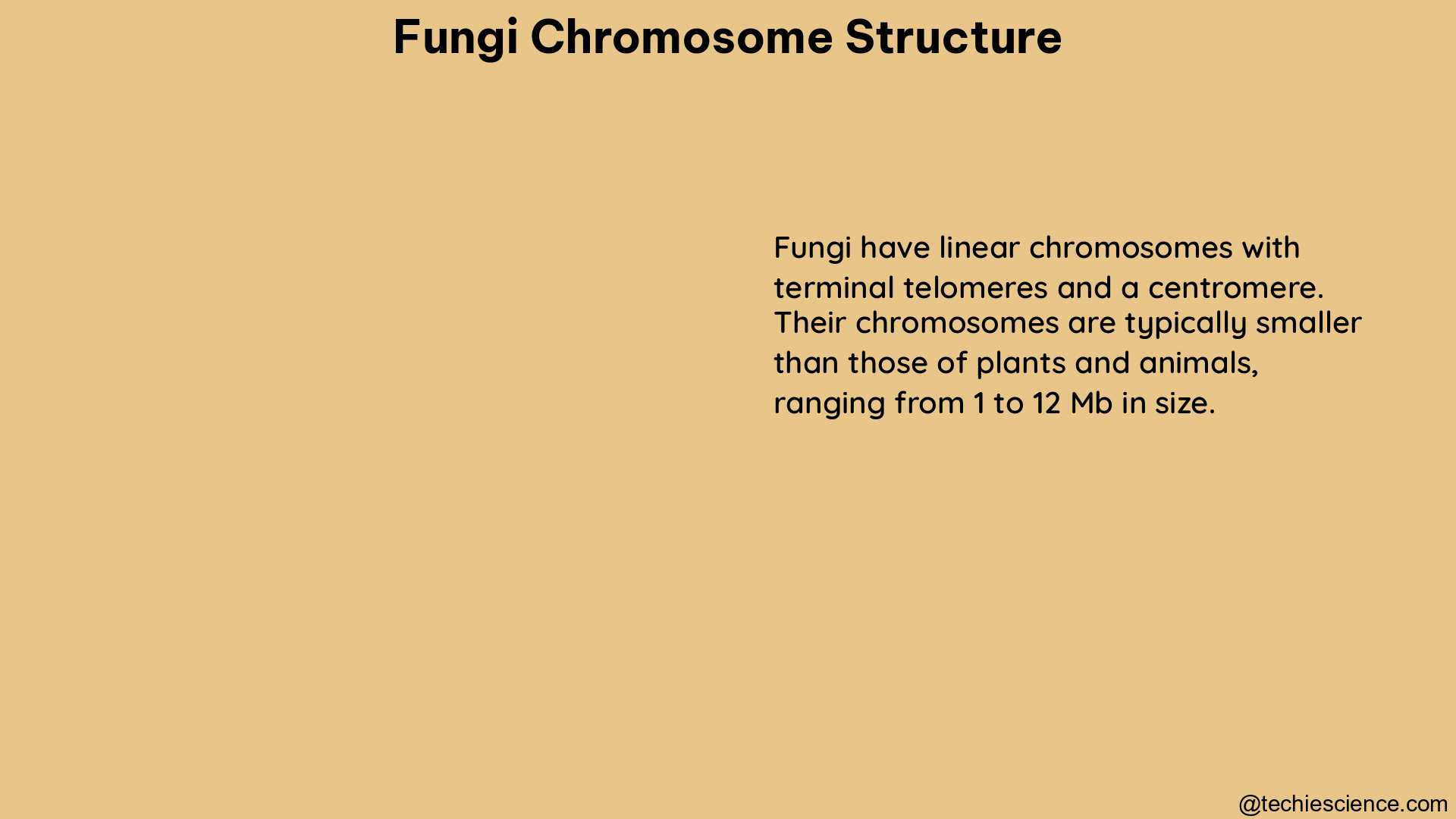
Fungi have relatively simple gene structures, which facilitate accurate gene prediction. The coding sequence length of genes in fungi averages between 1.3 and 2.5 kb. Centromeres in fungi are fragile sites that are often involved in karyotype variability and drug resistance. In most eukaryotes, centromeres are defined by specific DNA sequences, but in fungi, they are not well-defined and are often identified by their characteristic cytological features.
Fungal Genome Evolution and Chromosome Structure
-
Small-scale Genomic Changes: Research on fungal genome evolution has revealed that small-scale changes such as hypermutation, mobilization of various genetic elements, allelic transmission, and rapidly evolving regions can significantly impact the evolution of fungi.
-
Advances in Sequencing Technologies: Advances in long-range whole-genome sequencing technologies and the generation of telomere-to-telomere assemblies have allowed researchers to characterize the evolution of ploidy and chromosome structure, particularly in highly repetitive regions, with high accuracy and confidence.
-
Genome Composition Variability: Comparative genome analyses of eukaryotic pathogens including fungi and oomycetes have revealed extensive variability in genome composition. The genome size and ploidy level of pathogenic fungi and oomycetes can vary significantly between individuals of the same species.
-
Factors Influencing Genome Variability: Differences in genome composition can be attributed to the dynamics of transposable elements, chromosome instability, and genome compartmentalization.
-
Accessory Genome Compartments: Fungal genomes are known to contain accessory compartments that are thought to be relevant for rapid evolution of phytopathogens. Typically, these compartments contain a lower density of genes than the core genome, and have a higher content of repetitive elements.
Variant Discovery in Fungal Genomes
-
Genomic Variants: Population genomic datasets based on next generation sequencing (NGS) can be used to recover genomic variants such as single nucleotide polymorphisms (SNPs), insertions and deletions (indels), and structural variants (SVs).
-
Structural Variants: Structural variants include translocations, inversions, duplications (tandem or interspersed), deletions, and novel sequence insertions.
-
Variant Detection Frameworks:
- Reference-based Approach: Short read data generated from NGS is mapped on a reference genome to recover SNPs and short indel variants.
-
De novo Whole Genome Alignment (dnWGA): SNPs, indels, and SVs can be detected from whole genome alignments based on de novo assembled genomes.
-
Advantages of dnWGA: Whole genome alignment of de novo assembled genomes permits the joint analysis of genetic variation ranging from single nucleotide substitutions to large structural variation. SNPs can be called from WGAs with a precision similar to that of mapping-based approaches when sufficient sequencing coverage is achieved. The dnWGA approach also allowed the recovery of accessory chromosome fragments, which are shown to occur frequently in fungal genomes.
-
Challenges of dnWGA: In genomes with high frequencies of SVs and accessory regions, the use of dnWGA allows for reference-based mapping to be skipped entirely for variant discovery. However, current methods based on WGA are computationally more demanding than reference-based mapping approaches. As assembly algorithms are improving in quality and efficiency, fostered by the development of long-read sequencing technologies, whole genome alignment constitutes the next methodological challenge.
References
- Fungal Gene – an overview | ScienceDirect Topics
- The Host Adapted Fungal Pathogens of Pneumocystis Genus Utilize …
- Advances in understanding the evolution of fungal genome …
- On Variant Discovery in Genomes of Fungal Plant Pathogens
- Chromosome-Length Polymorphism in Fungi† – NCBI
Additional Resources
I am Ankita Chattopadhyay from Kharagpur. I have completed my B. Tech in Biotechnology from Amity University Kolkata. I am a Subject Matter Expert in Biotechnology. I have been keen in writing articles and also interested in Literature with having my writing published in a Biotech website and a book respectively. Along with these, I am also a Hodophile, a Cinephile and a foodie.