Enzymes and metabolism are the fundamental pillars of cellular physiology, orchestrating the intricate chemical reactions and energy production that sustain life. This comprehensive guide delves into the intricacies of enzymes, their regulation, and the pivotal role they play in the complex metabolic pathways that power cellular processes.
Understanding Enzymes: The Catalysts of Life
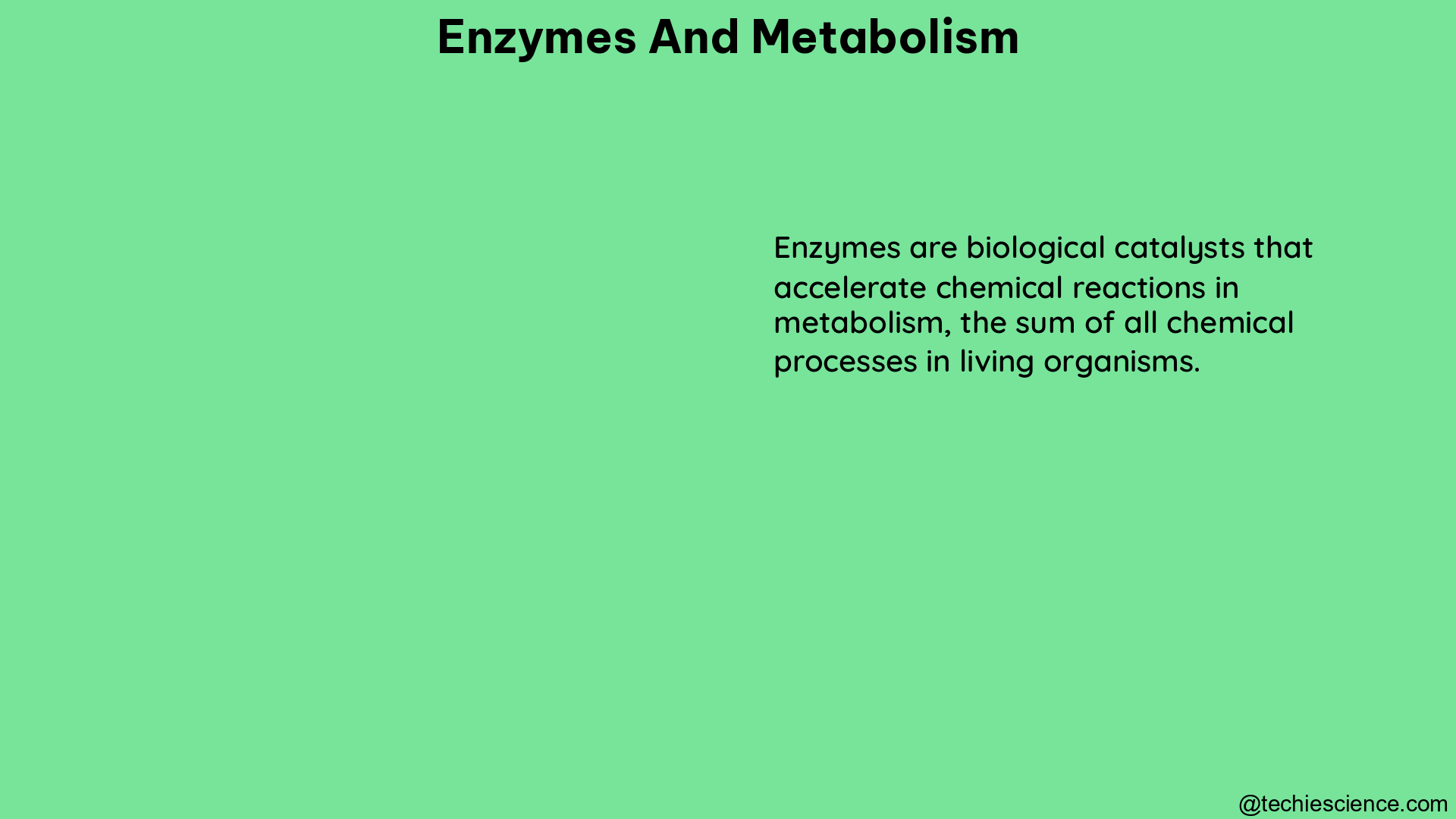
Enzymes are specialized proteins that act as catalysts, accelerating the rate of chemical reactions within cells. These remarkable biomolecules are responsible for catalyzing nearly all vital cellular processes, from energy production and nutrient breakdown to DNA replication and cellular signaling.
Enzyme Structure and Function
Enzymes are composed of intricate three-dimensional structures, with specific active sites that bind to and facilitate the transformation of substrates into products. The unique shape and chemical properties of an enzyme’s active site allow it to selectively bind and stabilize the transition state of a reaction, dramatically increasing the reaction rate.
Enzyme Kinetics and Regulation
The activity of enzymes is tightly regulated at multiple levels, including:
- Enzyme Concentration: The amount of an enzyme present in a cell or tissue can significantly impact its overall activity.
- Oligomeric State: Enzymes can exist in different oligomeric states, which can affect their catalytic efficiency and regulation.
- Allosteric Regulation: Binding of regulatory molecules to specific sites on an enzyme can induce conformational changes that modulate its activity.
- Covalent Modification: Post-translational modifications, such as phosphorylation or acetylation, can alter an enzyme’s structure and function.
- Compartmentalization: The localization of enzymes within specific cellular compartments can influence their accessibility to substrates and regulatory factors.
Understanding these regulatory mechanisms is crucial for deciphering the complex interplay between enzymes and their role in cellular metabolism.
Metabolic Mapping: Visualizing Enzyme Activity
Quantitative enzyme histochemistry and cytochemistry, also known as metabolic mapping, are powerful techniques used to study the activity and distribution of enzymes within cells and tissues.
Dehydrogenase Activity Mapping
Dehydrogenases are a class of enzymes that catalyze redox reactions, transferring electrons from one molecule (the substrate) to another (the cofactor). The activity of dehydrogenases can be visualized and quantified using metabolic mapping techniques.
- Tetrazolium Salt Reduction: Intact cells or cryostat tissue sections are incubated in a reaction medium containing the substrate, cofactors, and a tetrazolium salt. The dehydrogenase enzyme reduces the tetrazolium salt, forming a water-insoluble blue formazan precipitate at the site of enzyme activity.
- Quantification of Formazan Absorbance: The absorbance of the precipitated formazan can be measured using monochromatic light microscopy and image analysis, providing a direct quantitative measure of the local dehydrogenase activity.
Kinetic Parameter Determination
The quantitative nature of metabolic mapping enables researchers to determine in situ kinetic parameters, such as the maximal enzyme activity (Vmax) and the affinity of a substrate for an enzyme (Km). These parameters provide valuable insights into the regulation and efficiency of enzymes within their native cellular environment.
Enzyme Concentration and Reaction Rates
In vivo, the activity of enzymes can be influenced by their concentration within the cell or tissue. Interestingly, studies have shown that as enzyme concentration increases, the apparent enzymatic efficiency (Vmax/Km) can decrease, and the Km can increase.
Substrate Flux Limitation
Simulations have revealed that in situations with attenuated diffusion, the substrate flux can become the rate-limiting factor, rather than the enzyme concentration. This explains why reaction rates in vivo can sometimes be independent of enzyme concentrations, as the availability and transport of substrates to the enzyme active sites become the limiting factor.
Advances in Enzyme Activity Prediction
Recent developments in computational biology have enabled the prediction of Michaelis constants (Km) from structural features of enzymes. Deep learning algorithms have been trained on large datasets of enzyme structures and kinetic parameters, allowing for genome-scale predictions of Km values.
This powerful approach can provide valuable insights into the intrinsic properties of enzymes and their catalytic efficiency, further enhancing our understanding of cellular metabolism and the role of enzymes in various biological processes.
Conclusion
Enzymes and metabolism are the cornerstones of cellular physiology, driving the intricate chemical reactions and energy production that sustain life. By understanding the structure, function, and regulation of enzymes, as well as the techniques used to study their activity, we can gain deeper insights into the complex metabolic pathways that underpin cellular processes.
This comprehensive guide has explored the key concepts and advancements in the field of enzymes and metabolism, equipping you with the knowledge to navigate the intricacies of this fascinating area of biology. As research continues to unveil new discoveries, the understanding of enzymes and their role in metabolism will undoubtedly continue to evolve, shaping our understanding of the fundamental mechanisms that sustain life.
References:
- Molenaar RJ, Khurshed M, Hira VVV, Van Noorden CJF. Metabolic Mapping: Quantitative Enzyme Cytochemistry and Histochemistry to Determine the Activity of Dehydrogenases in Cells and Tissues. Frontiers in Physiology. 2018;9:561.
- Zotter A, Bäuerle F, Dey D, Kiss V, Schreiber G. Quantifying enzyme activity in living cells. Journal of Biological Chemistry. 2018;293(4):1303-1312.
- Kroll A, et al. Deep learning allows genome-scale prediction of Michaelis constants from structural features. PLoS Biology. 2021;19(10):e3001402.
- Cornish-Bowden A. Fundamentals of Enzyme Kinetics. Wiley-Blackwell; 2012.
- Nelson DL, Cox MM. Lehninger Principles of Biochemistry. W.H. Freeman; 2017.
- Berg JM, Tymoczko JL, Stryer L. Biochemistry. W.H. Freeman; 2015.

Hi….I am Shravanthi Vikram, I have completed my master’s in Bioinformatics and have 10 years of teaching experience in Biology.
Let’s connect through LinkedIn-